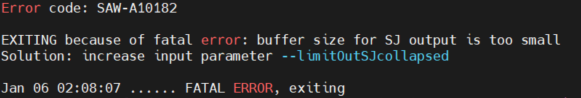
Pipeline | Code | Error type | Examples and error handling |
splitMask | SAW-A00001 | Parameters invalid or missing | e.g. "parameters error" Please check your input parameters. Some required parameters might be missed. e.g. "splitBcPos error, expected 1_24 or 2_25" Please check your input CID position is either 1_24 or 2_25. |
SAW-A00002 | File open failed | Please check the input file exists and has the correct access permission. |
SAW-A00003 | File parse failed | e.g. "only support .bin or .h5 file." Please check your input file is in the correct file format. |
SAW-A00004 | Other IO API error | e.g. "cannot write to file, /path/to/file" Please check your output directory path is an existing path. |
CIDCount | SAW-A00021 | Parameters invalid or missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A00022 | File open failed | e.g. "cannot open such file, /path/to/file" Please check the input file exists and has the correct access permission. |
SAW-A00023 | Failed to parser the file | Please check your input file is in the correct file format. |
SAW-A00024 | Other API error | Please check Appendix C or contact FAS/FBS for help. |
SAW-A00025 | Software exception | Please check Appendix C or contact FAS/FBS for help. |
mapping | SAW-A10101 | Parameters invalid or missing | e.g. "EXITING: FATAL INPUT ERROR: unrecognized parameter name "outSAMattribute" in input "Command-Line-Initial"" Please check the spelling of your argument and parameters. e.g. "please check the umi position and length" Please check the length of the reads in FQ1 are consistent with the parameters set for barcode length and umi length. For example, the length of the sum of barcode and umi set in bcPara file is 35 bp, but one of the reads in FQ1 is 30 bp, then you will see A10101 error code. Please check the parameters set in the bcPara file are consistent with your FQs. |
SAW-A10102 | File open failed | e.g. "Error, cannot open the file which be expected in gz or ascii format" Please check the file permission and file format. e.g. "barcodePositionMapFile does not exists: /path/to/mask" Please check the mask file exists and has the correct access permission. |
SAW-A10103 | File parse failed | e.g. "sequence and quality have different length" Please check the completeness of the reads in FQ. This issue may arise if the FQs are in incorrect format or the file was incompletely written or transferred. |
SAW-A10104 | Invalid data or data exception | Error data. Please check the file format and content. |
SAW-A10105 | File deletion failed | This error arose if "_STARtmp" directory failed to be deleted. Please check whether the program has completely finished according to the *.Log.progress.out or *.run.log file. |
SAW-A10106 | File IO failed | Please contact FAS/FBS for help. |
SAW-A10107 | Failure on APIs of system and libraries | Please contact FAS/FBS for help. |
SAW-A10108 | Software assert | Please contact FAS/FBS for help. |
SAW-A10109 | Software exception | Please contact FAS/FBS for help. |
SAW-A10110 | Allocate memory error | This error occurs when you run out of RAM. You may need to kill some jobs that use the same RAM or upgrade your hardware to get more RAM resources for your job. |
SAW-A10111 | Out of disk space | This error occurs when you store too many files on your hard disk. Please remove some files to free disk space. |
SAW-A10200 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A10201 | CID comparison rate is too low | This error usually arises because the CID information of the input FQs is not the same as the CID in the input mask file. Please use the correct SN-FQ pairs for mapping. |
SAW-A10202 | Fail to create the index for the BAM file | Please contact FAS/FBS for help. |
SAW-A10300 | Fail to load indexed reference | Please check the existence, access permission, and completeness for the indexed reference. |
SAW-A10301 | STAR parameter missing | Please check whether basic STAR parameters have been set. |
count | SAW-A20001 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A20002 | File open failed | Please check the input file that exists and has the correct access permission. |
SAW-A20003 | File parse failed | Please check your input BAM header is in the correct file format. |
SAW-A20004 | Allocate memory error | This error occurs when you run out of RAM. You may need to kill some jobs that use the same RAM or upgrade your hardware to get more RAM resources for your job. |
SAW-A20005 | File parse failed | e.g. "Found <number> gene names with their length exceeding 64 characters" Please check gene names in your input GTF/GFF file. |
SAW-A20101 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A20102 | File open failed | Please check the file that exists and has the correct access permission. |
SAW-A20103 | File parse failed | Please check your input file has a valid column number. |
SAW-A20105 | File parse failed | e.g. "Found <number> gene names with their length exceeding 64 characters" Please check gene names in your input GTF/GFF file. |
merge | SAW-A30001 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A30002 | File open failed | Please check the input file exists and has the correct access permission. |
SAW-A30003 | File parse failed | Please check your mask file is in the correct file format. Only support .h5/.bin mask file. |
SAW-A30004 | Allocate memory error | Please check whether the range of the coordinates is too large, or you have run out of RAM. You may need to kill some jobs that use the same RAM or upgrade your hardware to get more RAM resources for your job. |
SAW-A30005 | Fail to open input file | Fail to open input TXT file. Please check the file that exists and have the correct access permission. |
register & rapidRegister | SAW-A40001 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A40002 | File open failed | Please check the input file that exists and has the correct access permission. Or, please check whether a stitched panoramic TIFF (.tif or .tiff) image exists in the TAR.GZ. |
SAW-A40003 | File parse failed | Please check your input file is in the correct file format. The -v input gene expression matrix should be either a *tsv, barcode_gene_exp.txt, *.gem.gz, or *raw.gef. |
SAW-A40004 | Invalid data or data exception | Error data. Please check the file content. This error may arise because the -v input file is empty, the CZI file in TAR.GZ is invalid, or the QCPassFlag in IPR is 0. |
SAW-A40005 | Tissue segmentation error | Abnormal tissue segmentation reference score in image preprocessing. |
SAW-A40006 | IPR field missing | "Stitch/BGIStitch/StitchedGlobalLoc", does not exist. |
SAW-A40007 | Tiled image missing | Please check the uncompressed image folder has tiled images. |
SAW-A40008 | Insufficient GPU memory | GPU resources in the current node are insufficient. Please redeliver the task on an adequate one. |
SAW-A40009 | Abnormal chip SN prefix | Check "ImageInfo -> STOmicsChipSN" in.ipr where it offers the information of chip SN prefix. |
imageTools | SAW-A40401 | Parameter missing | Please check your imageTools merge input parameters. Some required parameters might be missed. |
SAW-A40402 | File open failed | Please check the imageTools merge input file that exists and has the correct access permission. |
SAW-A40405 | Invalid input | imageTools merge inputs of less than two images or more than three images.
|
SAW-A40406 | File pairing failed | Please check the imageTools merge input TIFF sizes are the same. Since the merge function is used for evaluating segmentation results, the input images are supposed to be the same in size and position (tissue position in the whole image). |
SAW-A40501 | Parameter missing | Please check your imageTools overlay input parameters. Some required parameters might be missed. |
SAW-A40502 | File open failed | Please check the imageTools overlay input file that exists and has the correct access permission. |
SAW-A40504 | Invalid data or data exception | Error data. Please check the file content. This error may arise because the -c input IPR file does not include Stitch/TransformTemplate or Register/MatrixTemplate information. |
SAW-A40601 | Parameter missing | Please check your imageTools img2rpi input parameters. Some required parameters might be missed. |
SAW-A40602 | File open failed | Please check the imageTools img2rpi input file that exists and has the correct access permission. |
SAW-A40605 | Invalid input | imageTools img2rpi input -i and -g have different length. These two inputs are supposed to be paired.
|
SAW-A40701 | Parameter missing | Please check your imageTools ipr2img input parameters. Some required parameters might be missed. |
SAW-A40702 | File open failed | Please check the imageTools ipr2imginput file that exists and has the correct access permission. Or, please check whether a stitched panoramic TIFF (.tif or .tiff) image exists in the TAR.GZ. |
SAW-A40703 | File parse failed | Please check your imageTools ipr2img input file is in the ImageQC/ImageStudio output TAR.GZ format. |
SAW-A40704 | Invalid data or data exception | Error data. Please check the imageTools ipr2img input IPR file content. This error may arise because the CZI file in TAR.GZ is invalid, or the image has not either automatically or manually registered with the expression matrix. The second circumstance can be confirmed from IPR by checking whether the StereoResepSwitch/register is TRUE (has not performed automatic registration) or the ManualState/register is FALSE (has not performed manual registration). The third possible reason is the StereoResepSwitch/tissueseg is TRUE, therefore cannot output cell segmentation result. |
SAW-A40706 | File pairing failed | Please check the shape of registered tissue segmentation images stored in theimageTools ipr2img input IPR are the same. Or please contact FAS/FBS for help. |
tissueCut | SAW-A50001 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A50002 | File open failed | Please check the file that exists and has the correct access permission. |
SAW-A50003 | File parse failed | Please check your input file is in the correct file format. |
SAW-A50004 | Fail to create output file | Fail to create output file. Please check your writing permission of the output directory. |
SAW-A50005 | Fail to write TIFF | Fail to write a TIFF file. |
SAW-A50006 | h5AttrWrite error | Please check the H5 file attributes. |
SAW-A50007 | h5DatasetWrite error | Please check the H5 file dataset. |
SAW-A50008 | Fail to create TIFF | Please check the access permission to write a TIFF image. |
SAW-A50009 | Different sizes between TIFF and GEF | Please check the sizes of the TIFF image and GEF (gene expression matrix) respectively. Make sure they are the same size. |
SAW-A50010 | Allocate memory error | This error occurs when you run out of RAM. You may need to kill some jobs that use the same RAM or upgrade your hardware to get more RAM resources for your job. |
cellCut | SAW-A60001 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A60002 | File open failed | Please check the file that exists and has the correct access permission. |
SAW-A60003 | File parse failed | The file does not contain correct information. Please check the file format. |
SAW-A60110 | Program version error | Please check your output GEF version. Your input GEF version might be too old. |
SAW-A60111 | Call process error | e.g. "Please call freeRestriction first, or call restrictRegion function before restrictGene." This error arose because the invocation flow order was messed up. Please modify your invocation flow as prompted. |
SAW-A60120 | Invalid data or data exception | Error data. Please check the file content. |
SAW-A60121 | File information missing | Failed to read the file. Please check whether the file is damaged. |
SAW-A60122 | File pairing failed | Please check the TIFF mask size is consistent with the size of expression matrix. Since the mask has been registered with the expression matrix, their sizes are supposed to be the same. |
SAW-A60130 | Fail to create output file | Fail to create output H5 file. Please check your writing permission of the output directory or contact FAS/FBS for help. |
SAW-A60140 | Allocate memory error | This error occurs when you run out of RAM. You may need to kill some jobs that use the same RAM or upgrade your hardware to get more RAM resources for your job. |
SAW-A60150 | Dimensions of gene expression matrix did not match | Please contact FAS/FBS for help. |
spatialCluster | SAW-A70001 | Parameter missing | e.g. "-i or --gef_file is missing" Please check your input parameters. Some required parameters might be missed. |
SAW-A70002 | File open failed | e.g. “cannot access /path/to/file: No such file or directory.” Please check the file that exists and has the correct access permission. |
SAW-A70005 | Value error | e.g. "The bin size is out of range, please check the range of gef binsize is in [1,10,20,50,100,200,500]." Please reset the bin size as prompted. e.g. "Gene number less than 3000, please check your gef file" Please check the content of your GEF file, and make sure there are at least 3000 genes for clustering. |
cellCluster | SAW-A70101 | Parameter missing | e.g. "-i or --gef_file is missing" Please check your input parameters. Some required parameters might be missed. |
SAW-A70102 | File open failed | e.g. “cannot access /path/to/file: No such file or directory.” Please check the file that exists and has the correct access permission. |
SAW-A70105 | Value error | e.g. "The bin size is out of range, please check the range of gef binsize is in [1,10,20,50,100,200,500]." Please reset the bin size as prompted. e.g. "Gene number less than 3000, please check your gef file" Please check the content of your GEF file, and make sure there are at least 3000 genes for clustering. |
saturation | SAW-A80001 | Parameter missing | e.g. "-i is missing." Please check your input parameters. Some required parameters might be missed. |
SAW-A80002 | File open failed | Please check the file that exists and has the correct access permission. |
SAW-A80003 | File parse failed | Invalid GEF file. Please check the file format. |
SAW-A80004 | Invalid data or data exception | e.g. "no data left after filter by coordinates." Please check the file content. |
SAW-A80005 | Invalid data or data exception | e.g. "total map reads is 0, please check file format from --bcstat" Please check the file content of the input file as prompted. |
SAW-A80006 | File pairing failed | e.g. "map reads less than annotated reads." Please check the input mapping statistical report and the count statistical report are from the same analysis. |
SAW-A80007 | Plot error | Please contact FAS/FBS for help. PATH environment may not have python3. |
SAW-A80008 | Fail to create output | Fail to generate saturation file, please contact FAS/FBS for help. |
report | SAW-A90001 | Parameter missing | e.g. "-m or --barcodeMapStat is missing." Please check your input parameter as prompted. Some required parameters might be missed. |
SAW-A90002 | File open failed | e.g. "cannot access *: No such file or directory." Please check the file that exists and has the correct access permission. |
SAW-A90003 | File parse failed | JSON file format error. This error may arise because the input statistics files were not generated in the same SAW version as report. Or, the input mapping file prefix can not be parsed. Please contact FAS/FBS for help. |
SAW-A90004 | Invalid data or data exception | e.g."information loss: fail to find 'bin_[size]' or 'ssDNA' in '*.rpi'." Please check the file content. |
SAW-A90005 | Fail to create output file | Fail to create output file. Please check your writing permission of the output directory. |
manualRegister | SAW-A40801 | Parameter missing | Please check your input parameters. Some required parameters might be missed. |
SAW-A40802 | File open failed | Please check the file that exists and has the correct access permission. Or, please check whether the pre-registered image fov_stitched_transformed.tif exists in the input directory. |
SAW-A40803 | File parse failed | Please check your input file is in the correct file format. The -v input gene expression matrix should be either a *tsv, barcode_gene_exp.txt, *.gem.gz, or *raw.gef. |
SAW-A40804 | Invalid data or data exception | Error data. Please check the file content. This error may arise because the -v input file is empty. The second possible reason is that the gene expression matrix information in the IPR Register module (MatrixShape, Xstart, Ystart) does not match with the input GEF file (minX, minY, maxX, maxY), because the manual registration has to be processed on the identical matrix. |
cellCorrect | SAW-A13001 | GEF parse failed | e.g. "-i parameter is invalid, please check your input." Please check the path or file format of GEF. |
SAW-A13002 | TIFF parse failed | e.g. "-m parameter is invalid, please check your input." Please check the path or size of TIFF image. |
SAW-A13003 | Fail to create output | e.g. "-o parameter is invalid, please check your input." Please check the access permission to write files or whether the output directory exists. |
SAW-A13004 | Invalid adjusting distance | e.g. "-d parameter exceeds the range of adjusting distance." Please check whether the adjusting distance is in a proper and reasonable range. |
lasso | SAW-A00031 | Parameters invalid or missing | e.g. "-i/-o/-m/-n is missing" Please check your input parameters. Some required parameters might be missed. |
SAW-A00032 | File open failed | e.g. "cannot access *: No such file or directory." Please check the file that exists and has the correct access permission. |
SAW-A00033 | File parse failed | e.g. "file type error: * is not GEF/GEOJSON file." Invalid file. Please check the file format. |