1. mapping
1.1 CID匹配和过滤统计结果
bash
$ cat /path/to/output/01.mapping/E100026571_L01_trim_read_1.CIDMap.stat
...
unique_CID_in_mask: 645784920
unique_CID_in_fq: 78021796
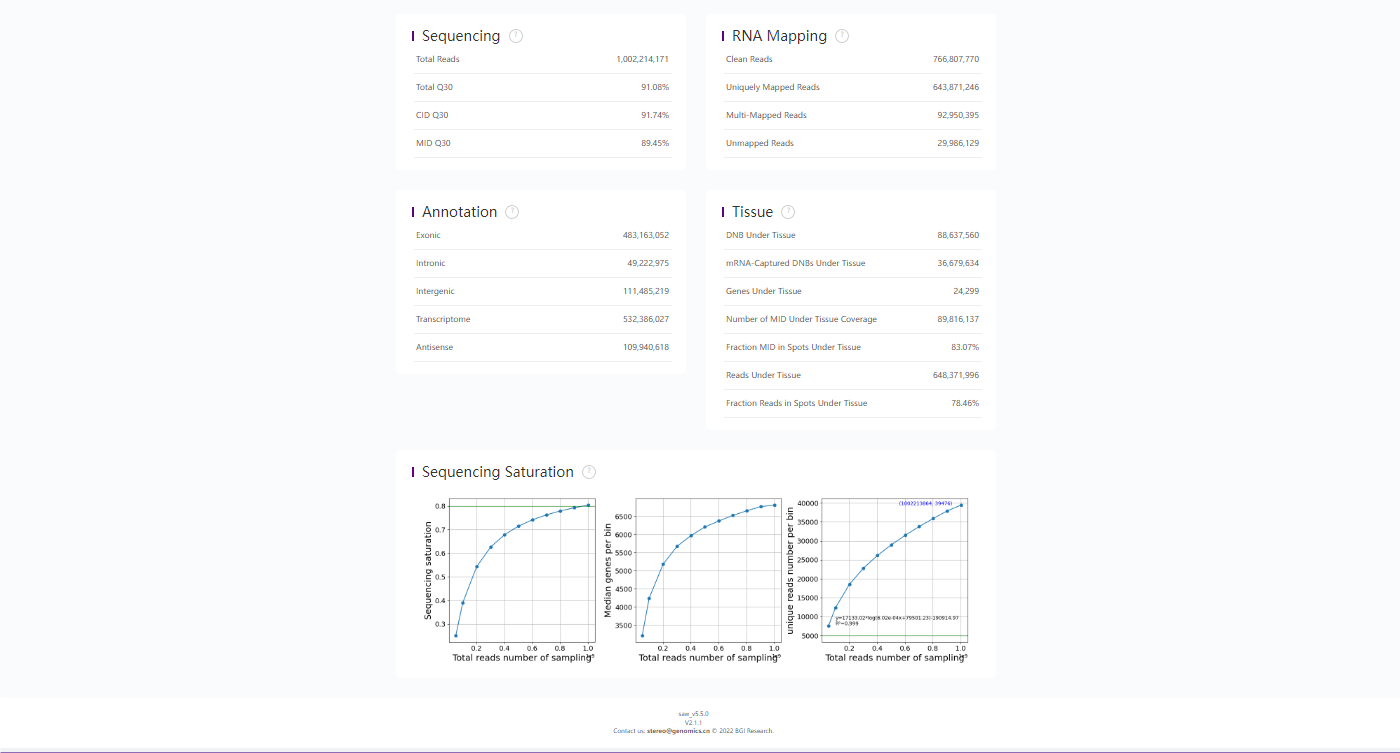
total_reads: 1002214171
CID_with_N_reads: 8031 0.00 %
CID_mapped_reads: 845113606 84.32 %
CID_exactly_mapped_reads: 698277934 69.67 %
CID_mapped_reads_with_mismatch: 146827641 14.65 %
discarded_MID_reads: 8451158 0.84 %
MID_with_N_reads: 13339 0.00 %
MID_with_polyA_reads: 13044 0.00 %
MID_with_low_quality_base_reads: 8424775 0.84 %
reads_with_dnb: 45281336 4.52 %
reads_with_adapter: 8136836 0.81 %
short_reads_filtered_with_polyA: 14191622 1.42 %
reads_with_polyA: 105849530 10.56 %
reads_with_rRNA: 0 0.00 %
Q10_bases_in_seq: 99.47 %
Q20_bases_in_seq: 97.12 %
Q30_bases_in_seq: 91.08 %
Q10_bases_in_MID: 99.26 %
Q20_bases_in_MID: 96.32 %
Q30_bases_in_MID: 89.45 %
Q10_bases_in_CID: 99.54 %
Q20_bases_in_CID: 97.49 %
Q30_bases_in_CID: 91.74 %
1.2 参考基因组比对统计
bash
$ cat /path/to/output/01.mapping/E100026571_L01_trim_read_1.Log.final.out
...
Number of input reads | 769052654
Average input read length | 95
UNIQUE READS:
Uniquely mapped reads number | 645319822
Uniquely mapped reads % | 83.91%
Average mapped length | 95.14
Number of splices: Total | 67634124
Number of splices: Annotated (sjdb) | 65711163
Number of splices: GT/AG | 66445242
Number of splices: GC/AG | 457783
Number of splices: AT/AC | 41573
Number of splices: Non-canonical | 689526
Mismatch rate per base, % | 0.50%
Deletion rate per base | 0.07%
Deletion average length | 3.91
Insertion rate per base | 0.03%
Insertion average length | 1.25
MULTI-MAPPING READS:
Number of reads mapped to multiple loci | 87828411
% of reads mapped to multiple loci | 11.42%
Number of reads mapped to too many loci | 5333051
% of reads mapped to too many loci | 0.69%
UNMAPPED READS:
Number of reads unmapped: too many mismatches | 0
% of reads unmapped: too many mismatches | 0.00%
Number of reads unmapped: too short | 29365488
% of reads unmapped: too short | 3.82%
Number of reads unmapped: other | 1205882
% of reads unmapped: other | 0.16%
CHIMERIC READS:
Number of chimeric reads | 0
% of chimeric reads | 0.00%
1.3 mapping BAM示例
bash
$ samtools view
/path/to/output/01.mapping/E100026571_L01_trim_read_1.Aligned.sortedByCoord.out.bam | head -2
E100026571L1C007R00303973559 256 1 3000644 3 100M * 0 0
GCCTCATTGTGCCCCATATGTTTGCCTATGTTGTGGACTTATTTTCATTAAACTTTAAAACATCTTTAATTTTTTTCTTTATTTCATCATTGACCAAGCT
-FCA9D?GFFD<-DF;EG,G?
NH:i:2 HI:i:2 AS:i:94 nM:i:2 Cx:i:8839 Cy:i:7539 UR:Z:120CF
E100026571L1C003R03702347721 0 1 3001778 255 100M * 0 0
GTATGACATCTGTCCAGGATCTTCTAGCTTTCATAGTCTCTGGTGAGAAGTCTGGAGTAATTCTAATAGGCCTGCATTTATATGTTACTTGACCTTTTTC
EEFEDFFEFFFFEFFFFEC@EFFFFDFFEEFFEFFFFCFCEFFAFBFCED??FGBEFFDC:FFFDCFAF4FAFFDFFDG?DFBD.F@FECA/FEDEFFAA
NH:i:1 HI:i:1 AS:i:92 nM:i:3 Cx:i:12136 Cy:i:14034 UR:Z:C0808
2. merge
2.1 CID对应reads数列表示例
bash
$ head /path/to/output/02.merge/SS200000135TL_D1.merge.barcodeReadsCount.txt
7127 18002 48
4348 19028 1
14130 8635 1
7618 14537 24
4912 10945 5
16783 12914 1
15539 8177 1
9288 8082 14
7274 16533 59
9087 10657 10
3. count
3.1 MID过滤和基因注释结果统计
$ cat /path/to/output/03.count/SS200000135TL_D1.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam.summary.stat
## FILTER & DEDUPLICATION METRICS
TOTAL_READS PASS_FILTER ANNOTATED_READS UNIQUE_READS FAIL_FILTER_RATE FAIL_ANNOTATE_RATE DUPLICATION_RATE
733148233 645319822 533743961 108363128 11.98 17.29 79.70
## ANNOTATION METRICS
TOTAL_READS MAP EXONIC INTRONIC INTERGENIC TRANSCRIPTOME ANTISENSE
645319822 645319822 484480416 49263545 111575861 533743961 110185975
100.0 100.0 75.1 7.6 17.3 82.7 17.1
3.2 注释结果BAM示例
$ samtools view /path/to/output/03.count/SS200000135TL_D1.Aligned.sortedByCoord.out.merge.q10.dedup.target.bam | head -2
E100026571L1C007R00303973559 768 1 3000644 3 100M * 0 0 GCCTCATTGTGCCCCATATGTTTGCCTATGTTGTGGACTTATTTTCATTAAACTTTAAAACATCTTTAATTTTTTTCTTTATTTCATCATTGACCAAGCT -FCA9D?GFFD<-D<CGFEGD-DG*FGFDFBE;E(9BGGE38FFFG9GG;0?GGFGB?E@G:GGG3GF79F0GGDG?G<D>F;EG,G?<<CD4>G=>B+C NH:i:2 HI:i:1 AS:i:94 nM:i:2 Cx:i:8839 Cy:i:7539 UR:Z:120CF
E100026571L1C003R03702347721 0 1 3001778 255 100M * 0 0 GTATGACATCTGTCCAGGATCTTCTAGCTTTCATAGTCTCTGGTGAGAAGTCTGGAGTAATTCTAATAGGCCTGCATTTATATGTTACTTGACCTTTTTC EEFEDFFEFFFFEFFFFEC@EFFFFDFFEEFFEFFFFCFCEFFAFBFCED??FGBEFFDC:FFFDCFAF4FAFFDFFDG?DFBD.F@FECA/FEDEFFAA NH:i:1 HI:i:1 AS:i:92 nM:i:3 Cx:i:12136 Cy:i:14034 UR:Z:C0808 XF:i:2
3.3 count基因表达文件示例
bash
$ h5dump -n /path/to/output/03.count/SS200000135TL_D1.raw.gef
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/03.count/SS200000135TL_D1.raw.gef | head -15
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_U32LE "x";
H5T_STD_U32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 76210618 ) / ( 76210618 ) }
DATA {
(0): {
636,
12671,
2
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/03.count/SS200000135TL_D1.raw.gef | head -20
HDF5 "/path/to/output/03.count/SS200000135TL_D1.raw.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 24670 ) / ( 24670 ) }
DATA {
(0): {
"0610005C13Rik",
0,
45
},
(1): {
3.4 count抽样文件
bash
$ head -8 /path/to/output/03.count/SS200000135TL_D1_raw_barcode_gene_exp.txt
x y geneIndex MIDIndex readCount
9602 7705 10551 611723 2
4888 10392 10551 665954 4
8901 7096 10551 881671 1
8901 7096 10551 357383 20
7397 18783 10551 355789 1
9155 13032 10551 297666 1
9155 13032 10551 298690 1
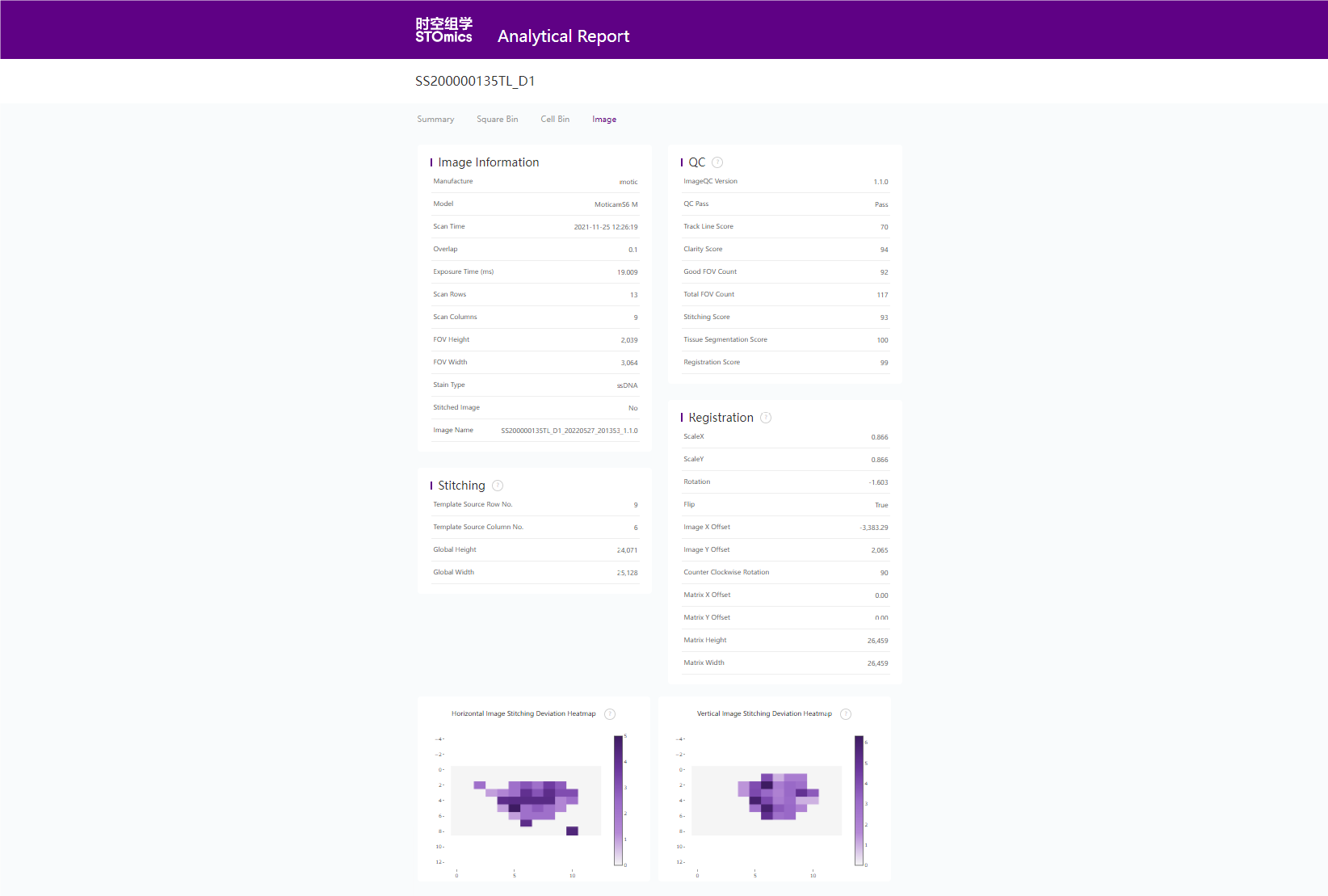
4. register and imageTools
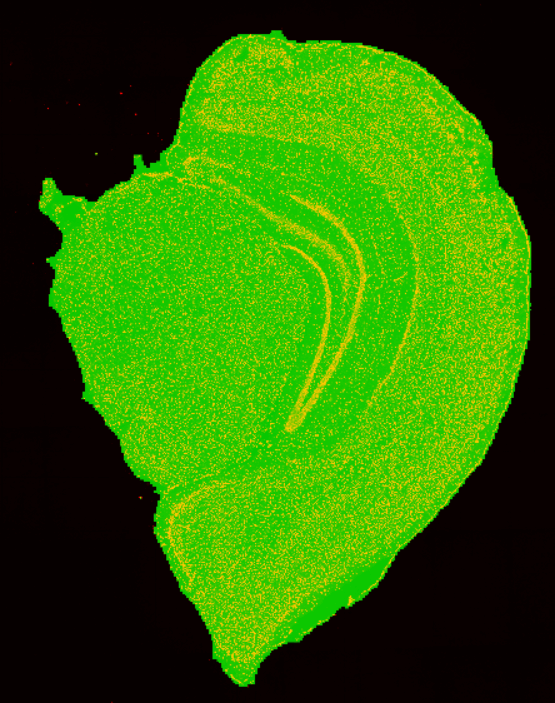
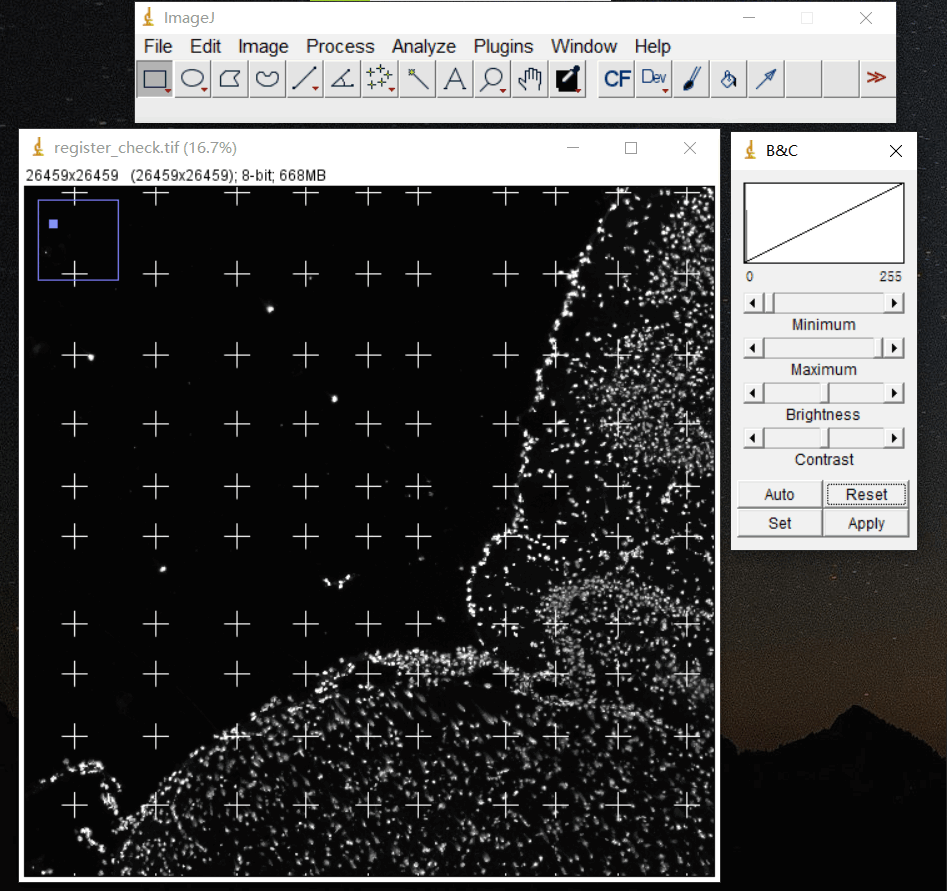
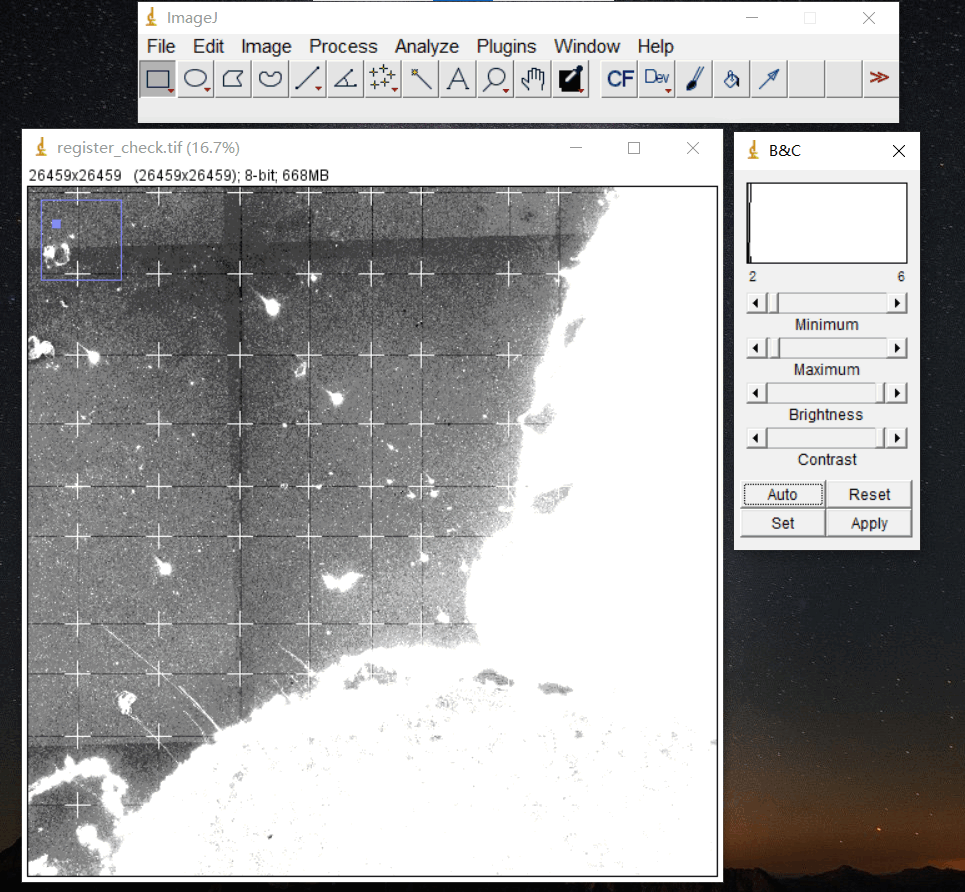
4.1 配准图
文件: /path/to/output/04.register/ssDNA_fov_stitched_transformed.tif和/path/to/output/04.register/ssDNA_SS200000135TL_D1_regist.tif
./path/to/output/04.register/ssDNA_fov_stitched_transformed.tif
/path/to/output/04.register/ssDNA_SS200000135TL_D1_regist.tif

4.2 图像处理过程记录文件
bash
$ h5dump -n /path/to/output/04.register/SS200000135TL_D1_20230822_144400_3.0.0.ipr
HDF5 "/path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr" {
FILE_CONTENTS {
group /
group /ManualState
dataset /Preview
group /StereoResepSwitch
group /ssDNA
group /ssDNA/CellSeg
dataset /ssDNA/CellSeg/CellMask
dataset /ssDNA/CellSeg/CellSegTrace
group /ssDNA/ImageInfo
dataset /ssDNA/ImageInfo/RGBScale
group /ssDNA/QCInfo
group /ssDNA/QCInfo/CrossPoints
dataset /ssDNA/QCInfo/CrossPoints/0_0
...
dataset /ssDNA/QCInfo/CrossPoints/9_8
group /ssDNA/Register
dataset /ssDNA/Register/MatrixTemplate
group /ssDNA/Stitch
group /ssDNA/Stitch/ScopeStitch
dataset /ssDNA/Stitch/ScopeStitch/GlobalLoc
dataset /ssDNA/Stitch/ScopeStitch/ScopeHorizontalJitter
dataset /ssDNA/Stitch/ScopeStitch/ScopeJitterDiff
dataset /ssDNA/Stitch/ScopeStitch/ScopeVerticalJitter
group /ssDNA/Stitch/StitchEval
dataset /ssDNA/Stitch/StitchEval/GlobalDeviation
dataset /ssDNA/Stitch/StitchEval/StitchEvalH
dataset /ssDNA/Stitch/StitchEval/StitchEvalV
dataset /ssDNA/Stitch/TemplatePoint
dataset /ssDNA/Stitch/TransformTemplate
group /ssDNA/TissueSeg
dataset /ssDNA/TissueSeg/TissueMask
}
}
$ h5dump -A /path/to/output/04.register/SS200000135TL_D1_20230822_144400_3.0.0.ipr | head -20
HDF5 "/path/to/output/04.register/SS200000135TL_D1_20220527_201353_1.1.0.ipr" {
GROUP "/" {
ATTRIBUTE "IPRVersion" {
DATATYPE H5T_STRING {
STRSIZE H5T_VARIABLE;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_UTF8;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "0.2.0"
}
}
GROUP "ManualState" {
ATTRIBUTE "calibration" {
DATATYPE H5T_ENUM {
H5T_STD_I8LE;
"FALSE" 0;
"TRUE" 1;
4.3 ImageTools merge
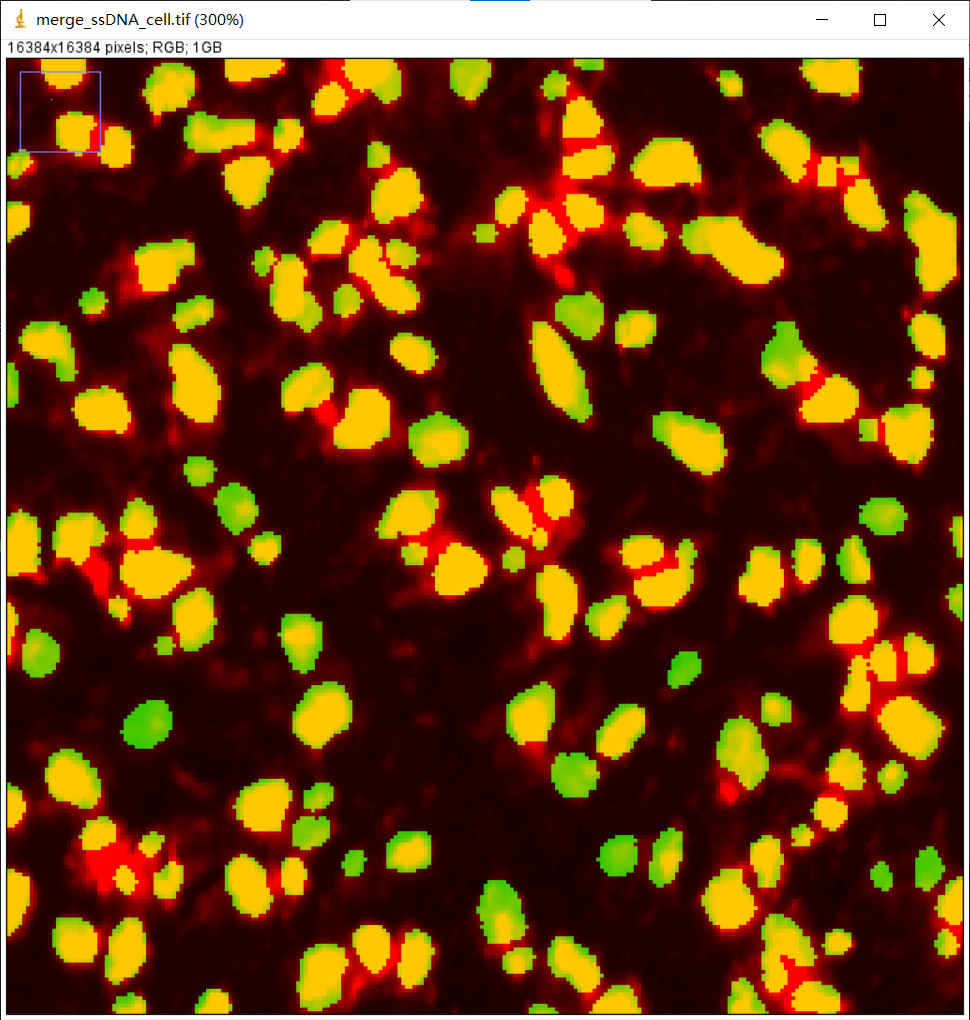
显微镜图像 ssDNA_SS200000135TL_D1_regist.tif和组织分割二值化掩膜文件ssDNA_SS200000135TL_D1_tissue_cut.tif 进行融合,来检查组织分割的结果。
显微图像ssDNA_SS200000135TL_D1_regist.tif 和细胞分割文件ssDNA_SS200000135TL_D1_mask.tif 进行部分融合,来检查细胞分割的结果。
4.4 ImageTools overlay
在 ssDNA_fov_stitched_transformed.tif 文件上叠加拼接模板来检查拼接的效果。
在 ssDNA_SS200000135TL_D1_regist.tif文件上叠加配准模板来检查配准的效果。
5. tissueCut
5.1 组织覆盖区域统计分析
bash
$ cat /path/to/output/05.tissuecut/tissuecut.stat
# Tissue Statistic Analysis with Stain ImageContour_area: 87086375
Number_of_DNB_under_tissue: 36521212
Ratio: 41.94%
Total_gene_type: 24289
MID_counts: 89679129
Fraction_MID_in_spots_under_tissue: 82.76%
Reads_under_tissue: 709807297
Fraction_reads_in_spots_under_tissue: 83.99%
binSize=1
Mean_reads_per_spot: 15.10
Median_reads_per_spot: 9.00
Mean_gene_type_per_spot: 1.71
Median_gene_type_per_spot: 1
Mean_Umi_per_spot: 2.46
Median_Umi_per_spot: 2
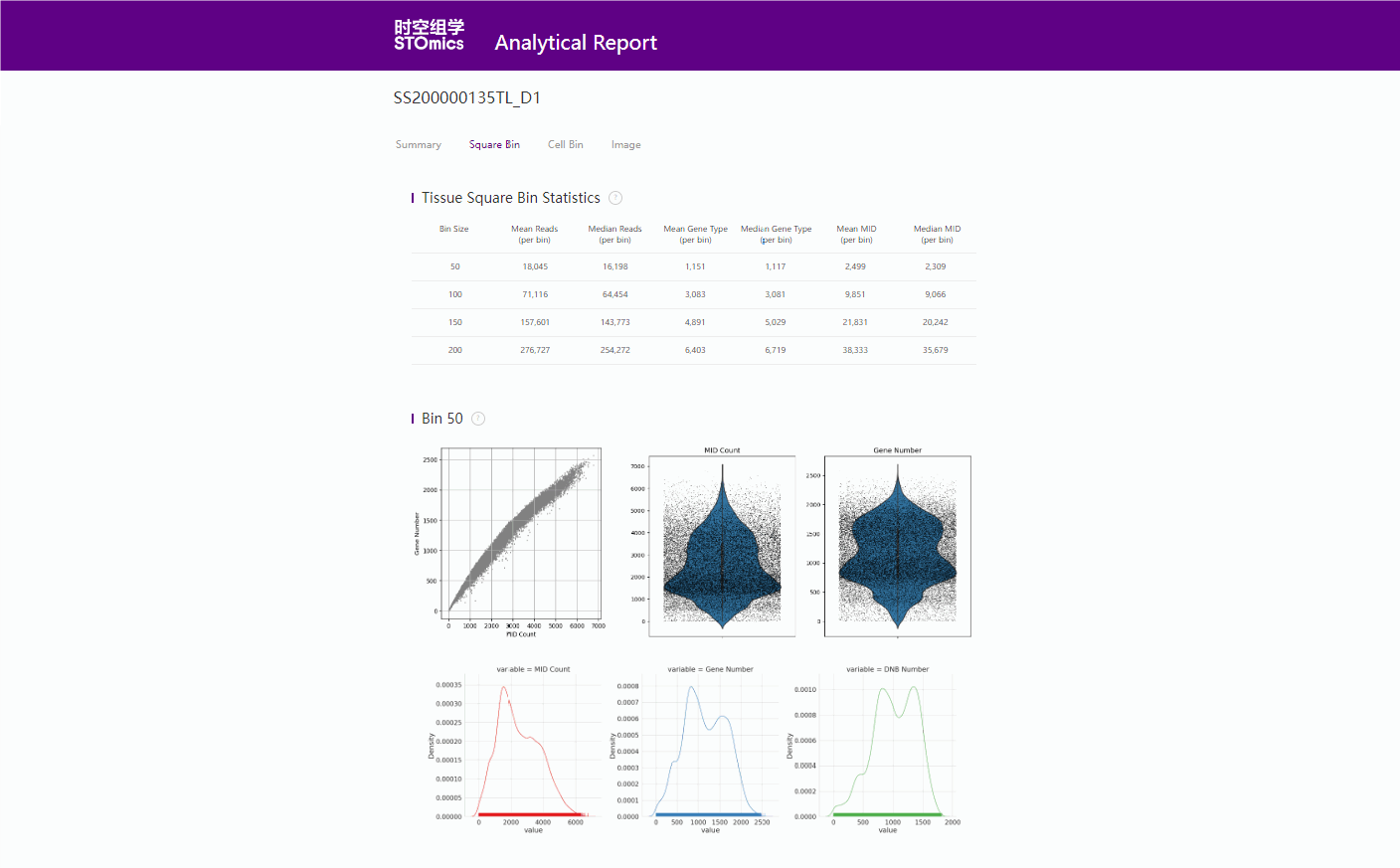
binSize=20
Mean_reads_per_spot: 3241.11
Median_reads_per_spot: 2782.00
Mean_gene_type_per_spot: 241.08
Median_gene_type_per_spot: 227
Mean_Umi_per_spot: 409.56
Median_Umi_per_spot: 370
binSize=50
Mean_reads_per_spot: 20054.45
Median_reads_per_spot: 18285.00
Mean_gene_type_per_spot: 1165.56
Median_gene_type_per_spot: 1133
Mean_Umi_per_spot: 2534.10
Median_Umi_per_spot: 2346
binSize=100
Mean_reads_per_spot: 78867.48
Median_reads_per_spot: 72545.00
Mean_gene_type_per_spot: 3110.83
Median_gene_type_per_spot: 3117
Mean_Umi_per_spot: 9964.35
Median_Umi_per_spot: 9205
binSize=150
Mean_reads_per_spot: 174614.34
Median_reads_per_spot: 162073.00
Mean_gene_type_per_spot: 4926.51
Median_gene_type_per_spot: 5065
Mean_Umi_per_spot: 22066.71
Median_Umi_per_spot: 20430
binSize=200
Mean_reads_per_spot: 305687.91
Median_reads_per_spot: 285723.00
Mean_gene_type_per_spot: 6424.38
Median_gene_type_per_spot: 6747
Mean_Umi_per_spot: 38621.50
Median_Umi_per_spot: 36060
5.2 组织覆盖区域基因表达矩阵示例
bash
$ h5dump -n /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
}
}
$ h5dump -d /geneExp/bin1/expression /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef | head -15
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
DATASET "/geneExp/bin1/expression" {
DATATYPE H5T_COMPOUND {
H5T_STD_U32LE "x";
H5T_STD_U32LE "y";
H5T_STD_U8LE "count";
}
DATASPACE SIMPLE { ( 62542665 ) / ( 62542665 ) }
DATA {
(0): {
6148,
10906,
1
},
(1): {
$ h5dump -d /geneExp/bin1/gene /path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef | head -20
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.tissue.gef" {
DATASET "/geneExp/bin1/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "offset";
H5T_STD_U32LE "count";
}
DATASPACE SIMPLE { ( 24289 ) / ( 24289 ) }
DATA {
(0): {
"0610005C13Rik",
0,
24
},
(1): {
5.3 补全GEF的基因表达矩阵示例
bash
$ h5dump -n /path/to/output/02.count/SS200000135TL_D1.gef
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.gef" {
FILE_CONTENTS {
group /
group /geneExp
group /geneExp/bin1
dataset /geneExp/bin1/exon
dataset /geneExp/bin1/expression
dataset /geneExp/bin1/gene
group /geneExp/bin10
dataset /geneExp/bin10/exon
dataset /geneExp/bin10/expression
dataset /geneExp/bin10/gene
group /geneExp/bin100
dataset /geneExp/bin100/exon
dataset /geneExp/bin100/expression
dataset /geneExp/bin100/gene
group /geneExp/bin20
dataset /geneExp/bin20/exon
dataset /geneExp/bin20/expression
dataset /geneExp/bin20/gene
group /geneExp/bin200
dataset /geneExp/bin200/exon
dataset /geneExp/bin200/expression
dataset /geneExp/bin200/gene
group /geneExp/bin50
dataset /geneExp/bin50/exon
dataset /geneExp/bin50/expression
dataset /geneExp/bin50/gene
group /geneExp/bin500
dataset /geneExp/bin500/exon
dataset /geneExp/bin500/expression
dataset /geneExp/bin500/gene
group /stat
dataset /stat/gene
group /wholeExp
dataset /wholeExp/bin1
dataset /wholeExp/bin10
dataset /wholeExp/bin100
dataset /wholeExp/bin20
dataset /wholeExp/bin200
dataset /wholeExp/bin50
dataset /wholeExp/bin500
group /wholeExpExon
dataset /wholeExpExon/bin1
dataset /wholeExpExon/bin10
dataset /wholeExpExon/bin100
dataset /wholeExpExon/bin20
dataset /wholeExpExon/bin200
dataset /wholeExpExon/bin50
dataset /wholeExpExon/bin500
}
}
$ h5dump -d /stat/gene /path/to/output/05.tissuecut/SS200000135TL_D1.gef | head -20
HDF5 "/path/to/output/05.tissuecut/SS200000135TL_D1.gef" {
DATASET "/stat/gene" {
DATATYPE H5T_COMPOUND {
H5T_STRING {
STRSIZE 64;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
} "gene";
H5T_STD_U32LE "MIDcount";
H5T_IEEE_F32LE "E10";
}
DATASPACE SIMPLE { ( 24670 ) / ( 24670 ) }
DATA {
(0): {
"Gm42418",
5860952,
60.1028
},
(1): {
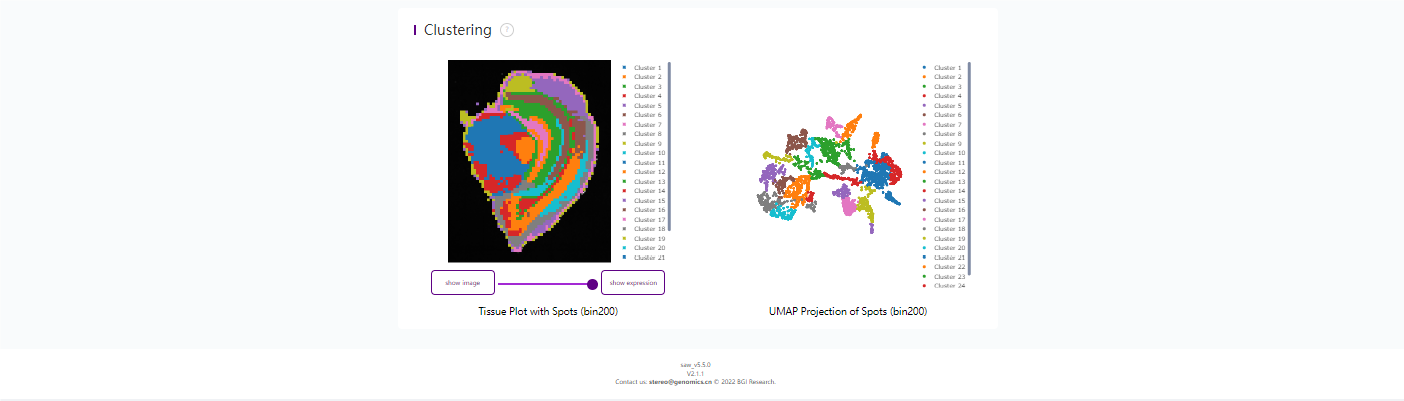
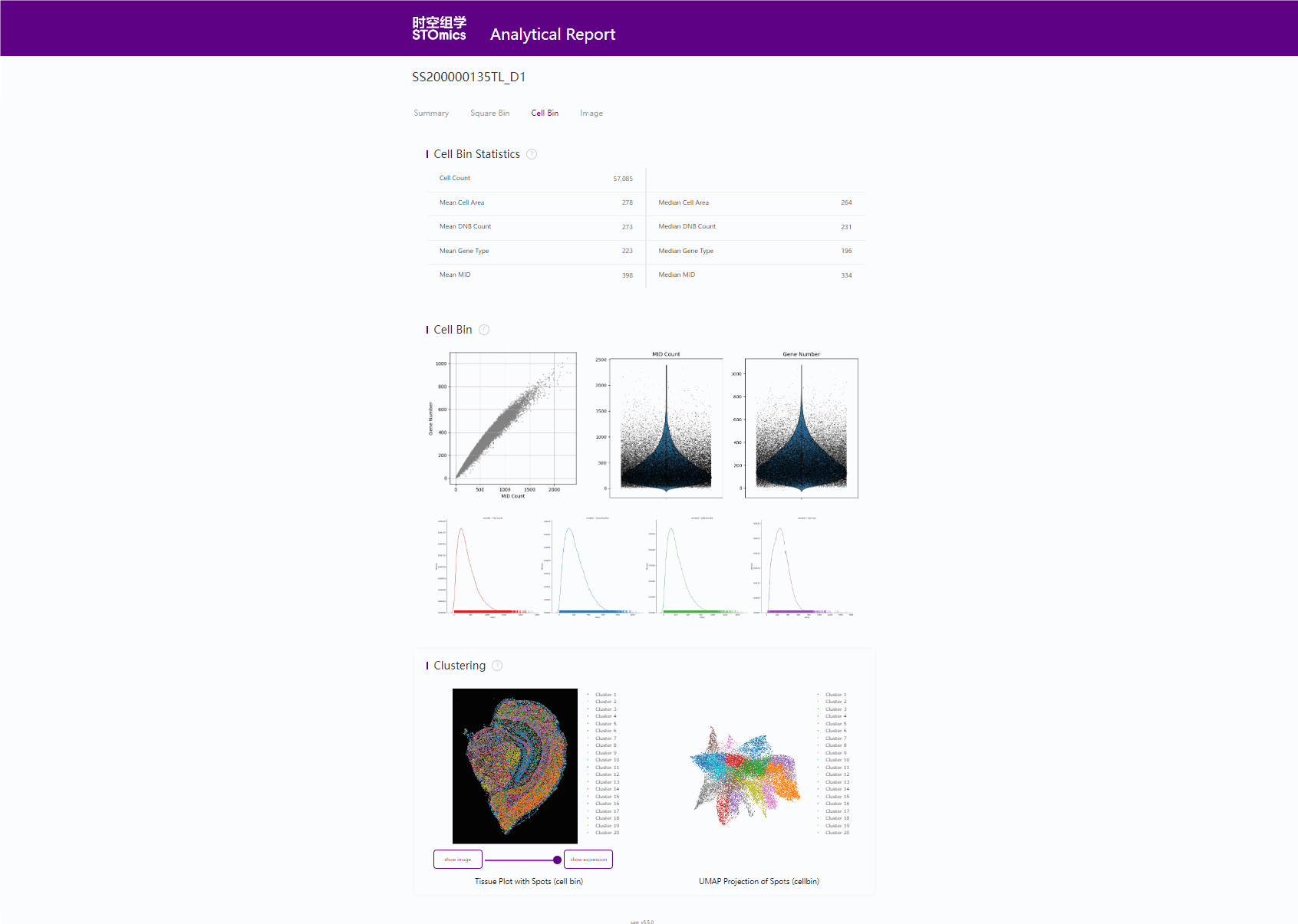
6. cellCut
6.1 cell bin 基因表达矩阵示例
bash
$ h5dump -n /path/to/output/051.cellcut/SS200000135TL_D1.cellbin.gef
HDF5 "/path/to/output/051.cellcut/SS200000135TL_D1.cellbin.gef" {
FILE_CONTENTS {
group /
group /cellBin
dataset /cellBin/blockIndex
dataset /cellBin/blockSize
dataset /cellBin/cell
dataset /cellBin/cellBorder
dataset /cellBin/cellExon
dataset /cellBin/cellExp
dataset /cellBin/cellExpExon
dataset /cellBin/cellTypeList
dataset /cellBin/gene
dataset /cellBin/geneExon
dataset /cellBin/geneExp
dataset /cellBin/geneExpExon
}
}
7. 测序饱和度文件示例
bash
$ cat /path/to/output/07.saturation/sequence_saturation.tsv
sample bar_x bar_y1 bar_y2 bar_umi bin_x bin_y1 bin_y2 bin_umi
0.05 26687198 0.250475 1 20002730 26687198 0.273744 3030 7041
0.1 53374396 0.389862 1 32565765 53374396 0.409615 4045 11464
0.2 106748792 0.542456 1 48842313 106748792 0.557064 4962 17194
0.3 160123200 0.625182 1 60017028 160123200 0.636707 5435 21128
0.4 213497584 0.677501 1 68852648 213497584 0.6871 5778 24238
0.5 266871984 0.713814 1 76374913 266871984 0.722081 6052 26886
0.6 320246400 0.740741 1 83026752 320246400 0.748035 6234 29228
0.7 373620768 0.761599 1 89071589 373620768 0.768155 6381 31356
0.8 426995168 0.778242 1 94689701 426995168 0.784222 6528 33334
0.9 480369568 0.79188 1 99974373 480369568 0.797395 6639 35194
1 533743961 0.803326 1 104973406 533743961 0.808462 6718 36641
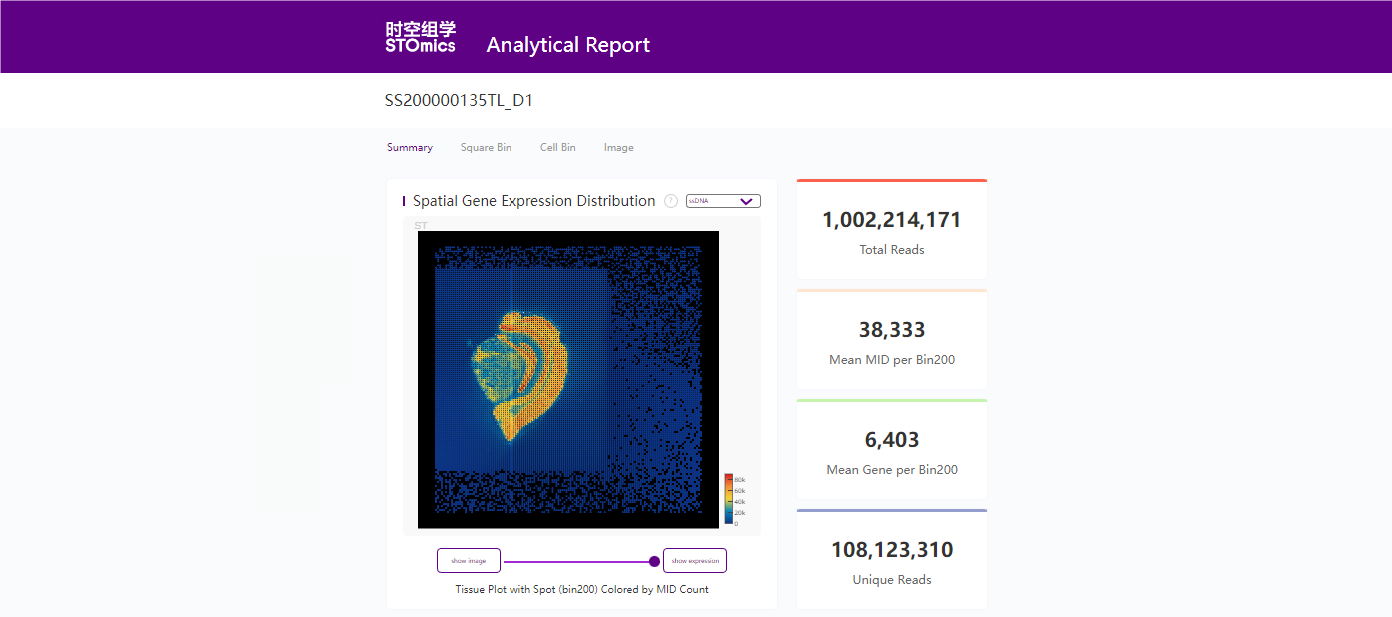
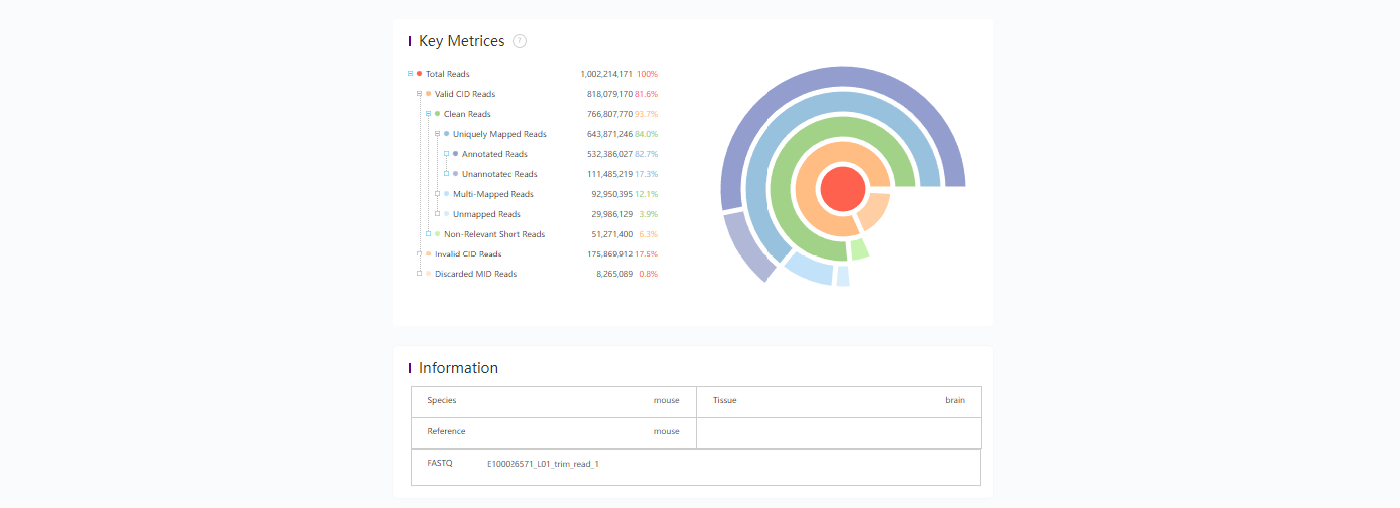
8. 报告
8.1 分析结果统计报告示例
bash
$ head /path/to/output/08.report/SS200000135TL_D1.statistics.json
{
"version": "version_v2",
"1.Filter_and_Map": {
"1.1.Adapter_Filter": [
{
"Sample_id": "E100026571_L01_trim_read_1",
"getCIDPositionMap_uniqCIDTypes": "645784920",
"total_reads": "1002214171",
"CID_withN_reads": "8031 (0.00 %)",
"mapped_reads": "845113606 (84.32 %)",
8.2 分析结果统计报告示例